Continental influx and pervasive matrilocality in Iron Age Britain – Nature.com
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain
the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in
Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles
and JavaScript.Advertisement
Nature
(2025)Cite this article
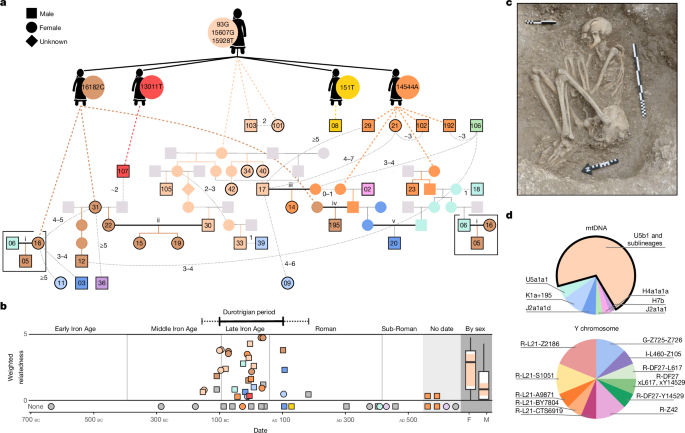
1499 AltmetricMetrics detailsRoman writers found the relative empowerment of Celtic women remarkable1. In southern Britain, the Late Iron Age Durotriges tribe often buried women with substantial grave goods2. Here we analyse 57 ancient genomes from Durotrigian burial sites and find an extended kin group centred around a single maternal lineage, with unrelated (presumably inward migrating) burials being predominantly male. Such a matrilocal pattern is undescribed in European prehistory, but when we compare mitochondrial haplotype variation among European archaeological sites spanning six millennia, British Iron Age cemeteries stand out as having marked reductions in diversity driven by the presence of dominant matrilines. Patterns of haplotype sharing reveal that British Iron Age populations form fine-grained geographical clusters with southern links extending across the channel to the continent. Indeed, whereas most of Britain shows majority genomic continuity from the Early Bronze Age to the Iron Age, this is markedly reduced in a southern coastal core region with persistent cross-channel cultural exchange3. This southern core has evidence of population influx in the Middle Bronze Age but also during the Iron Age. This is asynchronous with the rest of the island and points towards a staged, geographically granular absorption of continental influence, possibly including the acquisition of Celtic languages.The structure of a society is shaped by the residence patterns of its married couples4. Matrilocality, whereby partners predominantly reside with or near the wife’s parents, is relatively rare in modern ethnographic databases5,6, whereas patrilocality is by far the most common system. Furthermore, in most European Neolithic, Copper and Bronze Age sites with sufficient genomic and archaeological data, evidence of patrilocality and patriliny has been reported7,8,9,10,11,12,13.Despite being on the cusp of the historical era, little is known about the social structures of the Iron Age peoples of Britain. In the early centuries ad, Ptolemy described the locations of various ethne on the island with names of Celtic origin (Extended Data Fig. 1), and Caesar referred to civitates. These ambiguous terms are often translated as ‘tribes’, although the complexities of such group identities are not well understood. Interestingly, two of the earliest recorded British rulers were women, Cartimandua and Boudica, suggesting that both sexes could reach the highest political status. From Cartimandua’s 30-year reign of the Brigantes, a tribe covering much of northern England, we learn that women could inherit property, divorce and lead armies to great effect1. In the east of England, Boudica of the Iceni famously led an uprising that destroyed three Roman towns and challenged the authority of the imperial government14. Furthermore, Julius Caesar noted, in the mid-first century bc, that British women could take multiple husbands (De Bello Gallico). However, such social descriptions are seen as suspect, biased towards what would have seemed exotic to a Mediterranean audience that was immersed in a deeply patriarchal world1.The distributions of grave goods in multiple western European Celtic cemeteries have been interpreted as supporting high female status15. British archaeological evidence, however, is limited as Iron Age human remains are rare, with individuals perhaps predominantly cremated, excarnated or deposited in wetlands. The Durotriges tribe, who occupied the central southern English coast around 100 bc to ad 100, were one exception, depositing their dead in formal cemeteries of flexed inhumations (Fig. 1c and Supplementary Note 1). Interestingly, it is women who are more commonly associated with a greater number and diversity of prestige items in these burials, hinting at high status and perhaps a matrifocal society2.a, The best-fitting pedigree (for uncertainties, see Supplementary Note 4). Sampled individuals are outlined in black with WBK ID number and are coloured by mtDNA haplotype. The founding U5b1 + 16189 + @16192 female is shown at the top, with her four descendants with de novo mutations underneath. Further descendants are connected with dashed lines. Matings between descendants of the founding female are shown in bold, labelled i–v. Deduced relationships not fitted on the pedigree are shown with light-grey lines, with the estimated degree of relatedness. b, Weighted relatedness of each genome plotted versus the point carbon-14 date estimate (average 95% confidence range: 202 years). For each, the sum of their total number of biological kinship links (seventh degree or less) is shown, inversely weighted by the degree of the relationship. Individuals are coloured by mtDNA haplotype; grey indicates singleton haplogroups. The Durotrigian period (solid line) and the range of dates of family members (dashed line) are indicated. The summed relatedness is also shown in box plots (Tukey) by sex for individuals in the latter range; a significant difference between males (M) and females (F) is observed (Welch’s t-test, two-tailed, P = 0.029). The frequency of the dominant mtDNA lineage for each group is the proportion of each boxplot body in colour, which was also significantly different (two-tailed Fisher’s exact test, P = 0.02). c, A flexed inhumation excavated at WBK, typical of the Durotrigian cultural zone (photo credit: Bournemouth University). d, mtDNA and Y chromosome haplogroup frequencies for individuals with at least one genetic relative and sufficient Y chromosome coverage (Supplementary Table 9).The genomic variation of Iron Age Britons has been investigated16,17,18,19, but with limited data from single cemeteries that could clarify social customs relating to kinship and marriage. Genomic survey has contributed to debates on the spread of Celtic languages (Supplementary Note 1.6), with the Middle to Late Bronze Age identified as a candidate window for arrival based on the inference of large-scale migration to the island during this period, followed by substantial genetic isolation in the Iron Age17. However, the characterization of gene flow into Britain requires further refinement through haplotypic analysis and regional dissection. Here, we sequence 55 genomes from Durotrigian and other cemeteries at Winterborne Kingston (WBK), Dorset, along with two well-furnished female Durotrigian burials from Maiden Newton and Langton Herring2,20 (Supplementary Table 2 and Supplementary Note 1). These reveal a community characterized by female-line descent. When combined with data from other British Iron Age sites, our analyses find that matrilocality is widespread, reveal fine-scale genealogical networks that align with geographical boundaries and show a genomic footprint of Iron Age immigration on the south coast that is reflective of both contemporary Roman writing and archaeological datasets.Excavations at WBK in coastal southern England have revealed considerable evidence for settlement, spanning the later Bronze Age, around 1000 bc, to the post-Roman period, around ad 500, including several small Durotrigian-type cemeteries from the later Iron Age20 (Fig. 1b and Supplementary Note 1). Genomic data were retrieved for all 55 skeletal samples taken from the site (Supplementary Table 1), with 40 achieving a coverage high enough for genotype imputation and robust identification of genomic segments that were identical by descent (IBD) between individuals21 (>0.3×; Methods and Supplementary Note 3). This revealed WBK to be the burial ground of a large kin group during the Durotrigian period of the site’s usage (around 100 bc to ad 100; Fig. 1b), with 30 of 40 individuals possessing at least one relative of approximately the seventh degree or closer (Supplementary Table 10; see Supplementary Note 4.3 for exact criteria). An additional four low-coverage members of this kin group were identified through allele-matching analysis.Strikingly, more than two thirds (24/34) of the genetically identified kin belong to a rare lineage of mitochondrial haplogroup U5b1 (Fig. 1d) that has not been observed previously in ancient sampling and that has a frequency of only 3 × 10−5 in modern data22 (Supplementary Table 7 and Supplementary Note 2.4). The predominance of this single matriline is not skewed by an abundance of siblings, with only two pairs of sisters (all adults) observed (Fig. 1a). Additional downstream mutations distinguish four subclades in this haplogroup that are unique to WBK. Using one of the faster estimates of the mitochondrial DNA (mtDNA) mutation rate23 (4.72 × 10−7 mutations per site per generation), we estimate that at least 420 female births to lineage mothers would be required to result in this level of within-clade diversity (Supplementary Note 2.6), implying a long-term association between this haplotype and WBK. By contrast, we find that Y chromosome diversity is high (Fig. 1d and Supplementary Note 2.8), and runs of homozygosity (ROH) indicate that this was an outbreeding community (Supplementary Note 5.5). Theory, modelling and surveys of modern populations24,25 have demonstrated that such patterns are generated by matrilocal customs (that is, male-biased dispersal).To confirm matrilocality at WBK, we carried out two types of simulation (Supplementary Notes 2.10 and 4.5). First, we modelled different rates of male and female migration between demes in a population and estimated the resulting uniparental haplotype diversity (h) (Methods and Supplementary Note 2.2). These simulations indicated an outward female migration rate close to zero and a male rate between 0.15 and 1 per generation. Second, we simulated the distribution of autosomal and X chromosome kinship coefficients in a seven-generation pedigree whose members practised alternately (1) patrilocality, (2) matrilocality or (3) mixed residence. Again, the observed data are consistent with matrilocality (Supplementary Note 4.5). The earliest incidence of the dominant mtDNA lineage is in two second-degree relatives (346 to 51 calibrated (cal) bc), with the last observation in the Roman period (cal ad 31 to 212), when British Celtic societies underwent radical changes (Fig. 1b). Accordingly, the latest family member was buried following a new funerary rite of extended inhumation (WBK36; cal ad 82 to 316).We reconstructed the most parsimonious pedigree for the core kin group, which further confirms matrilocal traditions at WBK coupled with male mobility (Fig. 1a and Supplementary Note 4). We found only one patrilineal relationship greater than the first degree (WBK02 and WBK195), and we infer this to involve multiple partnerships with matriline women across generations. An adult woman (WBK31), her daughter (WBK22) and her adult granddaughters (WBK15 and WBK19) are all buried at the site, as well as an inferred matrilineal great-grandson (WBK12) of WBK31 through a different male partner. There is also one unusual case of a double relationship in our pedigree; from IBD segment length distributions, we can conclude that WBK17 is most likely the son of stepchildren whose parents’ marriage produced the sisters WBK34 and WBK40 (Supplementary Note 4.8).When we consider individuals dating to the Durotrigian period, we find that males show significantly lower levels of genetic relatedness with other individuals and are significantly overrepresented among non-matrilineage individuals (Fig. 1b). Six individuals, all male, show no detectable genetic connection to the WBK kin group (that is, they are not members of the dominant matriline and have no identified relatives), although they may still have been family members (for example, inward-migrating spouses or fostered children). Four of the six who were adult or adolescent at death were buried in typical Durotrigian fashion, three with grave goods comprising locally manufactured ceramic vessels, implying their integration in the community. When considering genetically related individuals, we find eight of ten family members who do not belong to the dominant mitochondrial haplogroup are male. We infer two marriages between these non-lineage men and lineage women, including the outlier WBK02 whose ancestry derives mainly from continental Europe (Extended Data Figs. 2 and 3).We note that the co-burial of spouses is not typical of a society with strict emphasis on matrilineal descent, in which men will frequently visit or even reside with their matrilineal kin and are often buried alongside them rather than with their wives26. Indeed, the integration of husbands into their wives’ households can place strain on matrilineal systems in which nephews inherit from their maternal uncles (the avunculate)27,28. For this reason, matrilocality is thought to be more stable when there is less property for male kin to control. It is associated with societies in which wealth is concentrated in the land, which is typically abundant and extensively farmed and owned by women, and in which men are often absent (for example, because of warfare)27,29,30.Interestingly, at WBK we infer five marriages in which both partners descend from the founding female (Fig. 1a), including three in which both members are direct descendants through the female line. However, these partners have no recent relatedness, as indicated by a lack of IBD sharing and lack of ROH in their offspring (Supplementary Note 5.5), and the matriline couples belong to different subclades. This suggests that the people of WBK had a deep knowledge of their own genealogies, which may have been used to guide marital arrangements among a pool of related groups in the local region. These patterns are consistent with modern matrilocal populations31 who typically show increased rates of local endogamy (for example, marriages of individuals from nearby villages or within the same village), which can allow men to retain influence in their natal group through geographical proximity.To place the WBK community in context, we searched for reduced mitochondrial diversity as a signature of matrilocal practice through space and time in Europe (Supplementary Note 2.2 and Supplementary Table 13). We considered 156 archaeological sites (first-degree relatives removed) spanning from the Neolithic to the Iron Age and observed six outlying communities with extremely low levels of diversity (Fig. 2 and Extended Data Fig. 1), all from the English Iron Age: Worlebury (Somerset), Bottle Knap (Dorset), Gravelly Guy (Oxfordshire), Trethellan Farm and Tregunnel (Cornwall) and Pocklington (Yorkshire). We further observed that the 11 lowest diversity estimates come from British Iron Age populations, as well as one English Middle to Late Bronze Age site. By contrast, Y chromosome diversity is high (Supplementary Table 18 and Supplementary Note 2.9), and patterns of ROH imply that these were relatively large outbreeding communities17 (Supplementary Note 5.5). At Pocklington17, the second-largest British cemetery sample in the dataset, 28 of 33 individuals belong to one of three dominant mtDNA haplogroups, which, in a manner akin to WBK, can be divided into subclades defined by private mutations. Here, the main period of burial activity was between 400 and 50 bc, but the first observation of a dominant matriline pre-dates this in the Early Iron Age (I11033; 717–395 cal bc; Supplementary Table 12).Trends in mtDNA haplotype diversity (h) for archaeological sites with two or more individuals after pruning of first-degree pairs. Haplotype diversity is calculated as the probability that two randomly selected haplotypes are different (Methods). In the bottom panels, the h value is plotted against the normalized number of relative pairs seen for each site (1, all pairs are genetic relatives; 0, no pairs are genetic relatives; Supplementary Note 5.3). The shaded area represents the 95% confidence interval around the fitted line. There is a strong negative correlation between mtDNA diversity and the number of relatives present for Iron Age sites (Pearson correlation coefficient, P = 0.001, r = −0.449), which is not observed in previous periods of prehistory. When each period is further split into continental and insular (UK and Ireland) individuals (diamonds and circles), we find that the only significant correlation observed is for the British Iron Age (Pearson correlation coefficient, P = 5.853 × 10−7, r = −0.717). The top panels show the geographical distribution of these h values for sites with evidence of burial guided by kinship (at least one pair of genetic relatives present). Of the total 156 sites considered, 13 sites are less diverse than WBK: 12 from Britain and 1 from a Celtic La Tène period cemetery (320–180 bc) in Hungary17. The sample sizes for the h value and normalized relative pair estimation for all sites are presented in Supplementary Table 13.These results provide strong evidence that longevous matrilocal communities were widespread across the island through the Iron Age and may even have their origins in the preceding Bronze Age period. Analyses of Bell Beaker and Early Bronze Age cemeteries in Britain and Germany have produced evidence of patrilocality and emphasis on patrilineal descent12,13,32, which, if reflective of the broader social organization of this period in Britain, raises the interesting possibility of a patrilocal society transitioning to matrilocality. This is a relatively rare occurrence in ethnographic surveys, although these may not be indicative of conditions throughout most of human history4,33.High mitochondrial diversity at a site may not solely reflect residence patterns but can also indicate an overall lack of biological relatedness among individuals; indeed, in Iron Age Britain, mtDNA diversity shows a significant (P = 5.85 × 10−7) inverse correlation with the normalized number of relative pairs identified using refinedIBD21 (Fig. 2). However, no similar reduction in mtDNA diversity is apparent for other prehistoric periods, despite the presence of multiple sites with high levels of biological relatedness (Fig. 2), implying that matrilocal practices were not widespread in Neolithic or Bronze Age Europe. By contrast, when we consider Y chromosome diversity in British Iron Age populations, no correlation with the number of relative pairs is identified (r = 0.06, P = 0.77; Supplementary Note 2.9).We found 30 instances of genetic relatives (more than 24 cM shared) between sites (most of which were between 2 km and 40 km distant), none of whom shared mtDNA haplotypes (Extended Data Fig. 4, Supplementary Note 5.6 and Supplementary Table 14). By contrast, 51% of within-site pairs share their mtDNA. For example, Dibbles Farm and Worlebury Hillfort on the Bristol Channel coast share eight relative pairs (30–55 cM IBD) and each site is dominated by a different matriline (Fig. 2 and Extended Data Fig. 1), suggesting that the movement was of male marriage partners. Similar patterns are seen in East Yorkshire, the region of the distinctive Iron Age Arras Culture associated with the Parisi tribe referenced by Ptolemy. We observe extreme levels of IBD sharing among all sites east of the River Derwent boundary, implying the existence of a cohesive social group in this territory (Extended Data Fig. 5). However, no shared mtDNA haplotype is observed between any of these East Yorkshire sites.To further characterize population structure in Iron Age Britain, we carried out Leiden clustering (Methods) on a weighted network graph of IBD sharing between archaeological sites (Fig. 3a and Supplementary Note 5.7). Consensus clusters were identified across 100 independent runs. These show clear geographical patterning; for example, subclusters in Scotland (greens), Yorkshire (blues), the Midlands (aquas) and the southwest (purples) all emerge. WBK is placed within a Dorset cluster (red), which maps onto the known distribution of later Iron Age ‘Durotrigian style’ coinage34,35 (Fig. 3c). Interestingly, several clusters encompass both continental and coastal British sites, pointing to cross-channel movements.a, The clusters are based on the consensus of 100 runs of the Leiden algorithm on a weighted graph of IBD shared between archaeological sites and show geographical integrity. Twelve major clusters (defining nodes marked with symbols) are labelled on the basis of geographical affiliations, with further substructure within clusters emphasized using different colour shades. The cross-channel clusters are highlighted with dashed lines joining nearest geographical neighbours across the channel. b, An interpolated map showing the distribution of British Bronze Age ancestry across Iron Age Britain, based on average values generated using ChromoPainter NNLS38 and SOURCEFIND37 approaches. The lowest values are seen along the south-central coast. Sites with less than 75% contribution are marked in black. c, A close-up showing most of the sites from the Dorset cluster (red circles) placed within the regional distribution of Durotriges coin finds. WBK is denoted by ‘W’. The distributions are plotted according to refs. 34,35. d, The EEF ancestry proportion through time for the channel core region of continental influence (blue; outlined with dashed line in b) shows a Late Iron Age increase not observed in the sample from the rest of England and Wales (black). The channel core zone is east of longitude −2.8° (western edge of the Durotrigian zone) and south of latitude 51.5° (River Thames). The period between 1000 and 875 bc (grey rectangle) has been previously associated with an increase in EEF ancestry in southern Britain17. This window is populated mostly by high-EEF samples from the channel core, whereas data points directly preceding this window are mostly from the peripheral regions that retained a lower level of EEF ancestry throughout the Middle Bronze Age (Extended Data Fig. 7 and Supplementary Note 6.2).Patterns of IBD segment sharing also reveal differences in population sizes across Britain and the continent (Extended Data Fig. 6). The south and east of England show markedly reduced levels of ROH and within-region IBD sharing, indicative of higher population densities and connectivity. These were very productive agricultural regions where the first proto-towns (oppida) of southern Britain emerged in the century before the Roman conquest of ad 43.An increase in continental ancestry components has been described for Iron Age genomes from the south of Britain (England and Wales)17 and has been interpreted as the result of large-scale movements into the island during and before the Late Bronze Age (around 1000 to 875 bc). This is detectable as a rise in Early European Farmer (EEF) ancestry (Supplementary Note 6.2). When we incorporate our data, we find a previously undetectable significant (Welch’s t-test, two-tailed, P = 0.0005) increase in EEF ancestry between the Early and Late Iron Age (from 39.7% ± 0.2% to 41.8% ± 0.5%), driven by genomes from southern regions along the central and eastern English Channel coast, including those from the Durotrigian territory (Fig. 3d and Supplementary Table 25). These regions emerged archaeologically as a core of unprecedented continental influence during the Middle Bronze Age, with cross-channel communities exhibiting parallel developments in disposal of the dead, settlement architecture and material culture over centuries, suggestive of high levels of population mobility3. Close cross-channel relations persisted throughout the Iron Age, when much of Britain seems to have developed a more regional and distinctively insular cultural footprint.When we split the genomic dataset into ‘channel core’ and ‘peripheral’ regions, we find that the rise of EEF ancestry during the Bronze Age was not a unitary process. Rather, the major increase in the channel core zone occurs across the Early to Middle Bronze Age, whereas a centuries-long lag is observed in the peripheral regions. For example, further regional division shows no increase in EEF ancestry in northern England from the Early Bronze Age until the Early Iron Age (around 750–400 bc; Extended Data Fig. 7).The impact of continental gene flow specific to the channel core zone is visible in principal-components analysis (PCA) of modern and ancient western Europeans (Extended Data Fig. 2), as well as patterns of haplotype copying from continental populations, characterized using ChromoPainter36 (Fig. 3b). We used SOURCEFIND37 to decompose the ancestry of Iron Age genomes into contributions from Early Bronze Age British and continental groups and further validated our results using an alternative approach of non-negative least squares38 (NNLS) with a different panel of surrogates (Methods and Supplementary Note 6.3). Overall, we estimate an average contribution of 73% (estimated by SOURCEFIND; NNLS estimate: 75%) from the British Early Bronze Age (2500 to 1500 cal bc) to the English and Welsh Iron Age population (800 bc to ad 50). Although this value is larger than the estimate of a previous study17, which inferred a 50% long-term replacement rate for the gene pool, it is in agreement with the reported dilution of British- and Irish-specific R1b-L21 haplogroup Y chromosomes by one quarter17.A sharp dip in Bronze Age continuity is seen along the channel coast (Fig. 3b and Extended Data Fig. 8). This is centred on Hampshire (SOURCEFIND estimate of 60%), a region traditionally associated with Belgic tribes that Caesar mentioned as having migrated from Gaul3. Both Hampshire and the neighbouring Durotrigian zone show independent and significant increases in EEF ancestry between the Early and Late Iron Age (Extended Data Fig. 7). Notably, the Durotrigian territory was home to a major port at Hengistbury Head, one of the focal points of intensifying cross-channel networks as Roman influence spread across Gaul39. With fewer samples for analysis, haplotypic data provide less resolution on fine-grained temporal trends but identify numerous genetic outliers in the Middle to Late Iron Age, all from the channel core region, which are not discernible when EEF ancestry alone is considered (Extended Data Fig. 3; see Supplementary Note 6.3 for further discussion of genetic outliers). These outliers include one of the most elaborate warrior burials known for Iron Age England (North Bersted on the channel coast; around 50 cal bc), which has been proposed, on the basis of isotopic signature and burial rite, to belong to a stream of cross-channel migrants, fuelled by Caesar’s conquest of Gaul40.Regional continuity is strongest in Scotland, estimated at 92%, with contributions preferentially coming from the Scottish Early Bronze Age population (Extended Data Fig. 8 and Supplementary Note 6.3). Large components of Early Bronze Age ancestry are also seen in northern England (88%) and the southwest (78%). Outside of Britain, a single Netherlands Late Iron Age genome also shows some evidence of population continuity, deriving its ancestry almost entirely from the Netherlands Bronze Age population in SOURCEFIND analysis. By contrast, French populations show a diversity of components, mainly from French and German sources, but with large minor components of Czech Iron Age ancestry in the east and Spanish Bronze Age ancestry in the south, highlighting France’s position as a crossroads in the Celtic-speaking world. We note one French outlier from the coastal site Urville-Nacqueville41, which faces Dorset across the English Channel and contains Durotrigian-style flexed burials in shallow oval graves. This individual has an estimated 72% contribution from the British Bronze Age, implying that gene flow occurred in both directions across the channel.The diverse geography of Britain lends itself to regionality, which manifests across archaeological periods3. In its Iron Age we characterize fine-grained geographical genetic structure, shaped by natural territorial boundaries such as rivers. The peripheral regions—including Scotland, Cornwall, Wales and northern England—show signatures of insularity. The southern channel core is an exception, showing reduced genomic continuity with the British Early Bronze Age, sites with cross-channel IBD affinities, indications of larger population size and individuals with outlying ancestries. In this region, we see a Middle to Late Iron Age spike in EEF ancestry, indicative of substantial cross-channel movements that match textual and archaeological evidence for an intensification of contact and exchange, driven, at least latterly, by Roman expansion into Gaul.The flow of genes across the channel through the Bronze and Iron Ages provides a wide window for the arrival of Celtic languages. Substantial components of continental ancestry are present in the channel core region by the Middle Bronze Age. However, it is probable that a second surge of EEF ancestry in the Iron Age would have influenced any version of insular Celtic already spoken in the channel region, and we note that the Celtic languages of southern Britain (Brittonic) and Gaul share a number of innovations not seen in more peripheral branches, such as the Goidelic languages of Ireland and Scotland42. Given the strong signatures of Early Bronze Age continuity in most British regions, any language introduction after this period would have probably been driven by a demographic minority, potentially an elite.It is possible that the pervasive matrilocal traditions of Iron Age Britain were also introduced from the continent, but, notably, reduced mtDNA diversity is pronounced in our peripheral populations (Fig. 2 and Extended Data Fig. 1). Matrilineal succession has previously been proposed for continental Celtic societies, on the basis of the discovery of a likely avuncular relationship between two ‘princely’ burials of the Hallstatt elites in Central Europe43. Matrilineal institutions may also have been present in the British Iron Age, given that social units based on unilineal descent are common in large agricultural societies that practise unilocal residence4. However, the burial of male spouses at WBK suggests that, if matrilineal descent groups existed in this society, they were limited in their function26. We note that in matrilocal societies with a weak avunculate, mother–daughter–sister relationships are generally given more emphasis, with women tending to enjoy relatively higher status and control over property27.Both matrilocality and matriliny are predicted by cultural factors that increase female involvement in subsistence labour and decrease paternity certainty28,29,44,45,46. External warfare can encourage both of these through male absence and has long been theorized to induce transitions to matrilocality through various mechanisms45,47,48, a hypothesis recently strengthened through quantitative modelling49. Matrilocality also predicts a history of migration into a new territory, which often is accompanied by frontier warfare4,45. The British Iron Age was debatably a time of high societal violence, indicated by the early proliferation of hillforts, weapons, human remains displaying violence-related injuries and instances of intergroup conflict recorded by Roman writers such as Julius Caesar and Tacitus50,51,52,53. Importantly, although matrilocality does not necessitate female political and social empowerment, it is strongly associated with these4,27,54,55,56 and resonates with Roman descriptions of Celtic women1. Although classical depictions of conquered peoples are often viewed with scepticism, we find here some truths in these writers’ appraisal of Iron Age Britain.We sampled 57 burials for DNA sequencing from three sites in Dorset2,20,57,58,59—WBK (n = 55), Langton Herring (n = 1) and Maiden Newton (n = 1). Petrous bones were preferentially sampled (n = 46), alongside tooth roots (n = 10) and a single phalanx. Sample processing took place in clean-room facilities dedicated to ancient DNA research at Trinity College Dublin. DNA extraction was carried out following various protocols60,61,62,63 detailed in Supplementary Table 4. DNA extracts were treated with USER enzyme to reduce post-mortem deamination lesions, and double-stranded libraries were created for Illumina sequencing61,64. Library aliquots were amplified using Accuprime Pfx Supermix (Life Technologies) with sample-specific index primers (Supplementary Table 5). Paired-end or single-end sequencing was carried out on MiSeq, HiSeq 2500 and NovaSeq 6000 platforms (Supplementary Table 5).Exact P7 index matches were required for demultiplexing, with up to two mismatches allowed in the P5 index for paired-end data. Adapters were removed from single-end data with cutadapt65 and from paired-end data with AdapterRemoval66. Paired-end reads with an overlap of 11 bp were collapsed. Singleton reads and collapsed reads that required quality trimming were discarded. Reads were mapped to GRCh37 with decoy contigs (hs37d5) using BWA software67 with non-default parameters -l 16500, -n 0.02 and -o 2. Reads were sorted with SAMtools68, polymerase chain reaction duplicates were removed with Picard Tools v.2.0.1 and indels were locally realigned using GATK software (v.3.7.0)69. Reads with a mapping quality below 25 and a read length below 34 bp were removed. Finally, we ‘soft-clipped’ the data by reducing the Phred quality scores of the two terminal base pairs at the 5′ and 3′ read ends to a score of 2. Comparative ancient genomic sequence data were downloaded and realigned from either unaligned FASTQ (when available) or BAM (aligned binary alignment map) files following the same pipeline (Supplementary Table 12).A detailed description of uniparental marker analysis is found in Supplementary Note 2. In brief, for mitochondrial haplotype calling, unfiltered read data aligned to GRCh37 were realigned to the Cambridge Reference Sequence for human mtDNA and subjected to the same downstream filters as described for GRCh37 alignments. Variants were called using BCFtools (v1.10.2)70, and the resulting VCF (variant call format) file was inputted into HaploGrep2 (ref. 71) to assign haplogroups based on Phylotree (Build 17)72. To estimate contamination, we calculated the fraction of minor alleles at HaploGrep-identified single-nucleotide variant sites present in the sample (Supplementary Table 7). Haplotype diversity (h) for archaeological sites was calculated as the probability that two randomly selected haplotypes were different73,74 (Supplementary Table 13). For Y chromosome haplotype calling, we relaxed several filters in our read processing pipeline: (1) we did not require an exact P7 index match; (2) we included singletons and collapsed reads that required quality trimming; (3) we filtered for a mapping quality above 20 and read length above 30 bp; and (4) we did not carry out soft-clipping. We used the Pileup tool from GATK (v.3.7.0)69 to extract base calls for positions in the International Society of Genetic Genealogy (ISOGG) database of Y chromosomal markers (version 15.73, 11 July 2020) and The Big Tree database (https://www.ytree.net/). Base calls below a quality of 30 were removed. The allelic state for each male sample at relevant markers was then assessed (Supplementary Table 9). Haplogroups used for within-site estimates of Y chromosome diversity in Britain are presented in Supplementary Table 18.We used pseudo-haploid genotypes for PCA and quantification of EEF ancestry. We used the Pileup tool from GATK software (v.3.7.0)69 to extract base calls over single-nucleotide polymorphism (SNP) sites in the 1,240k panel75 for relevant genomes and selected one base call at random (base quality >30) for each site to generate pseudo-haploid genotypes. We merged 1,240k genotypes for 534 Iron Age individuals17,18,19,41,61,76,77,78,79,80 with a dataset of 5,326 modern individuals from western Europe38,81 and, using approximately 266,000 sites common to both datasets, projected ancient genomes onto a PCA plot of modern variation using smartpca (version 16000) from EIGENSOFT82. We quantified EEF ancestry in British Iron Age genomes following a previously described procedure17. In brief, the qpAdm tool83, implemented in the ADMIXTOOLS2 R package, was used to model British Bronze and Iron Age genomes as a mixture of western hunter-gatherer, EEF and steppe pastoralist ancestries (Supplementary Tables 12 and 15). Whole-genome sequence data, rather than targeted SNP capture, were used for source and reference outgroup populations. Source populations61,76,79,84,85,86,87 were a set of Mesolithic individuals from northwest Europe (n = 13), Yamnaya pastoralists (n = 6) and Early Neolithic Europeans from central and southeastern Europe (n = 9). Reference populations79,84,88,89,90 were a set of Mesolithic individuals from Latvia and Romania (n = 6), Afanasievo pastoralists (n = 4), Anatolian Neolithic farmers (n = 11) and 10 modern-day Mbuti individuals from the Congo region of Africa91. Further information on PCA and qpAdm analyses is provided in Supplementary Note 6.We carried out genotype imputation on a dataset of 2,054 ancient individuals, including 42 individuals from the current study using GLIMPSE software92 (Supplementary Table 12). This included both whole-genome sequence (>0.1×) and targeted SNP capture (more than 300,000 calls across the 1,240k panel) datasets. After imputation, we further filtered for low-coverage individuals by extracting 1,240k panel positions and removing individuals for whom more than 40% of those positions had a genotype probability below 0.99. Stricter downstream filters were subsequently applied depending on the downstream analysis. To avoid any potential batch effects, we imputed each sample individually with GLIMPSE using the 1000 Genomes Project haplotype reference panel93. We used reference datasets and pipelines available on the software’s webpage (https://odelaneau.github.io/GLIMPSE/glimpse1/).Four datasets of GLIMPSE-imputed diploid genotypes (genotype probability >0.99) were subjected to IBD segment identification (Supplementary Table 12). To identify segments, each of the four datasets was subjected to further phasing and imputation using Beagle5 (ref. 94), followed by refinedIBD analysis (Supplementary Note 3). Different sets of variant sites were used as input into both Beagle5 and refinedIBD to test performance and maximize IBD segment retrieval. This resulted in 21 runs of refinedIBD in total, all carried out with default parameters. The outputted IBD segments were subsequently subjected to different merges and filters depending on the downstream application. Patterns of IBD segment sharing were characterized within (ROH) and between genomes, as well as within and between archaeological sites (Supplementary Note 5). We created a weighted graph of average IBD sharing between Iron Age sites in northwest Europe and performed hierarchical community detection using the Leiden algorithm95 implemented in the R package leidenAlg (v1.1.1)96. We ran the leiden.community function 100 times with different seeds and constructed a consensus tree from the output using the maximum clade credibility function available in the R package phangorn (v2.11.1)97.To reconstruct familial relationships at WBK, we used a combination of data types, including (1) uniparental markers; (2) autosomal coefficients of relatedness that were calculated using both allele-frequency-based methods and IBD segment sharing; (3) IBD1 and IBD2 segment numbers and lengths for genomes with more than 0.3× coverage, which were compared with distributions simulated using ped-sim98; (4) longest observed IBD segments within the genome; and (5) X chromosome IBD segment sharing. We determined the most likely genealogical relationships for pairs of relatives of first- to fourth-degree relatives (Supplementary Note 4), allowing us to construct the most parsimonious pedigree for the WBK kin group.We used a dataset of 697 individuals17,18,19,41,76,77,78,99,100,101,102,103,104,105,106 from the European Bronze Age to medieval period for ChromoPainter36 analysis (Supplementary Table 12). This dataset had been previously subjected to Beagle5 imputation and phasing. We extracted 1,240k SNP sites and rephased these using SHAPEIT2 (v2.r837)107. Two separate panels of surrogate individuals were then selected and ChromoPainter was used to generate co-ancestry matrices summarizing the amount of haplotypic donations between pairs of surrogates following recommended guidelines. One panel (n = 332) was then subjected to fineSTRUCTURE clustering using a previously described maximum concordance tree-building method38. This panel was used to paint a set of British Iron Age genomes, whose ancestry was then decomposed into contributions from the identified fineSTRUCTURE clusters (n = 17) using NNLS regression. The second panel (n = 307) was grouped into populations based on archaeological era and geographical location, rather than fineSTRUCTURE cluster, and contained only targeted SNP capture data. This panel was used to paint a larger set of British Middle to Late Bronze and Iron Age genomes, as well as Iron Age genomes from France and the Netherlands. Target populations included both whole-genome sequence and SNP capture data. Ancestry profiles were then generated using SOURCEFIND37. SOURCEFIND was run using 50,000 burn-in iterations followed by 200,000 sample iterations, thinning every 5,000 iterations. We set the expected number of surrogates used to form the target as two, with a total number of four surrogates allowed to form the target in each iteration. We carried out 50 independent runs of the above procedure and extracted the estimates with the highest posterior probability in each run. The average of these 50 estimates (weighted by posterior probability) was then calculated for each individual. This provided us with a set of ancestry proportions for each genome. We observed a strong correlation between SOURCEFIND and NNLS results with respect to British Bronze Age haplotype contributions. Further details can be found in Supplementary Note 6.3.The R package ggplot2 was used for figure generation (https://ggplot2.tidyverse.org). Maps were generated using the R packages maps (10.32614/CRAN.package.maps) and mapdata (10.32614/CRAN.package.mapdata). For Extended Data Fig. 4, the retired rgeos package and raster package were used, with data from the public Database of Global Administrative Areas.Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.Aligned sequence reads are available through the European Nucleotide Archive under accession number PRJEB81465. Other relevant data are available from the corresponding authors on reasonable request.This study made use of publicly available software, referenced throughout the main text and Supplementary Information.Allason-Jones, L. in A Companion to Women in the Ancient World (eds James, S. L. & Dillon, S.) Ch. 34, 467–477 (Blackwell, 2012).Russell, M., Smith, M., Cheetham, P., Evans, D. & Manley, H. The girl with the chariot medallion: a well-furnished, Late Iron Age Durotrigian burial from Langton Herring, Dorset. Archaeol. J. 176, 196–230 (2019).Article
Google Scholar
Cunliffe, B. Britain Begins (OUP Oxford, 2013).Ember, C. R., Droe, A. & Russell, D. in Explaining Human Culture (ed. Ember, C. R.) (Human Relations Area Files https://hraf.yale.edu/ehc/summaries/residence-and-kinship, accessed 01/10/2024).Murdock, G. P. et al. D-PLACE dataset derived from Murdock et al. 1999 ‘Ethnographic Atlas’ (v3.0). Zenodo https://doi.org/10.5281/zenodo.10177061 (2023).Kirby, K. R. et al. D-PLACE: a global database of cultural, linguistic and environmental diversity. PLoS ONE 11, e0158391 (2016).Article
PubMed
PubMed Central
MATH
Google Scholar
Chyleński, M. et al. Patrilocality and hunter-gatherer-related ancestry of populations in East-Central Europe during the Middle Bronze Age. Nat. Commun. 14, 4395 (2023).Article
ADS
PubMed
PubMed Central
MATH
Google Scholar
Fowler, C. et al. A high-resolution picture of kinship practices in an Early Neolithic tomb. Nature 601, 584–587 (2022).Article
ADS
CAS
PubMed
MATH
Google Scholar
Schroeder, H. et al. Unraveling ancestry, kinship, and violence in a Late Neolithic mass grave. Proc. Natl Acad. Sci. USA https://doi.org/10.1073/pnas.1820210116 (2019).Article
PubMed
PubMed Central
MATH
Google Scholar
Furtwängler, A. et al. Ancient genomes reveal social and genetic structure of Late Neolithic Switzerland. Nat. Commun. 11, 1915 (2020).Article
ADS
PubMed
PubMed Central
MATH
Google Scholar
Villalba-Mouco, V. et al. Kinship practices in the early state El Argar society from Bronze Age Iberia. Sci. Rep. 12, 22415 (2022).Article
ADS
CAS
PubMed
PubMed Central
Google Scholar
Mittnik, A. et al. Kinship-based social inequality in Bronze Age Europe. Science 366, 731–734 (2019).Article
ADS
CAS
PubMed
MATH
Google Scholar
Sjögren, K.-G. et al. Kinship and social organization in Copper Age Europe. A cross-disciplinary analysis of archaeology, DNA, isotopes, and anthropology from two Bell Beaker cemeteries. PLoS ONE 15, e0241278 (2020).Article
PubMed
PubMed Central
Google Scholar
Mackay, D. Echolands: A Journey in Search of Boudica (Hachette UK, 2023).Pope, R. Re-approaching Celts: origins, society, and social change. J. Archaeol. Res. 30, 1–67 (2022).Article
MATH
Google Scholar
Morez, A. et al. Imputed genomes and haplotype-based analyses of the Picts of early medieval Scotland reveal fine-scale relatedness between Iron Age, early medieval and the modern people of the UK. PLoS Genet. 19, e1010360 (2023).Article
CAS
PubMed
PubMed Central
Google Scholar
Patterson, N. et al. Large-scale migration into Britain during the Middle to Late Bronze Age. Nature 601, 588–594 (2022).Article
ADS
CAS
PubMed
MATH
Google Scholar
Martiniano, R. et al. Genomic signals of migration and continuity in Britain before the Anglo-Saxons. Nat. Commun. 7, 10326 (2016).Article
ADS
CAS
PubMed
PubMed Central
MATH
Google Scholar
Schiffels, S. et al. Iron Age and Anglo-Saxon genomes from East England reveal British migration history. Nat. Commun. 7, 10408 (2016).Article
ADS
CAS
PubMed
PubMed Central
MATH
Google Scholar
Russell, M. et al. The Durotriges Project 2016: an interim statement. Proc. Dorset Nat. Hist. Archeol. Soc. 138, 105–111 (2017).MATH
Google Scholar
Browning, B. L. & Browning, S. R. Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics 194, 459–471 (2013).Article
PubMed
PubMed Central
MATH
Google Scholar
MITOMAP. A human mitochondrial genome database. FOSWIKI http://www.mitomap.org (2023).Zaidi, A. A. et al. Bottleneck and selection in the germline and maternal age influence transmission of mitochondrial DNA in human pedigrees. Proc. Natl Acad. Sci. USA 116, 25172–25178 (2019).Article
ADS
CAS
PubMed
PubMed Central
MATH
Google Scholar
Lansing, J. S. et al. Kinship structures create persistent channels for language transmission. Proc. Natl Acad. Sci. USA 114, 12910–12915 (2017).Article
ADS
CAS
PubMed
PubMed Central
MATH
Google Scholar
Oota, H., Settheetham-Ishida, W., Tiwawech, D., Ishida, T. & Stoneking, M. Human mtDNA and Y-chromosome variation is correlated with matrilocal versus patrilocal residence. Nat. Genet. 29, 20–21 (2001).Article
CAS
PubMed
MATH
Google Scholar
Ensor, B. E., Irish, J. D. & Keegan, W. F. The bioarchaeology of kinship: proposed revisions to assumptions guiding interpretation. Curr. Anthropol. 58, 739–761 (2017).Article
Google Scholar
Fox, R. Kinship and Marriage: An Anthropological Perspective (Cambridge Univ. Press, 1984).Fortunato, L. The evolution of matrilineal kinship organization. Proc. Biol. Sci. 279, 4939–4945 (2012).PubMed
PubMed Central
MATH
Google Scholar
Mattison, S. M. Evolutionary contributions to solving the ‘matrilineal puzzle’: a test of Holden, Sear, and Mace’s model. Hum. Nat. 22, 64–88 (2011).Article
PubMed
MATH
Google Scholar
Surowiec, A., Snyder, K. T. & Creanza, N. A worldwide view of matriliny: using cross-cultural analyses to shed light on human kinship systems. Philos. Trans. R. Soc. Lond. B 374, 20180077 (2019).Article
MATH
Google Scholar
Ly, G. et al. From matrimonial practices to genetic diversity in Southeast Asian populations: the signature of the matrilineal puzzle. Philos. Trans. R. Soc. Lond. B 374, 20180434 (2019).Article
MATH
Google Scholar
Booth, T. J., Brück, J., Brace, S. & Barnes, I. Tales from the Supplementary Information: ancestry change in Chalcolithic–Early Bronze Age Britain was gradual with varied kinship organization. Cambr. Archaeol. J. 31, 379–400 (2021).Article
Google Scholar
Shenk, M. K., Begley, R. O., Nolin, D. A. & Swiatek, A. When does matriliny fail? The frequencies and causes of transitions to and from matriliny estimated from a de novo coding of a cross-cultural sample. Philos. Trans. R. Soc. Lond. B 374, 20190006 (2019).Article
Google Scholar
Papworth, M. The Search for the Durotriges: Dorset and the West Country in the Late Iron Age (History Press, 2011).Sellwood, L. in Aspects of the Iron Age in Central Southern Britain (eds Cunliffe, B. & Miles, D.) 191–204 (Oxford Univ. School of Archaeology, 1984).Lawson, D. J., Hellenthal, G., Myers, S. & Falush, D. Inference of population structure using dense haplotype data. PLoS Genet. 8, e1002453 (2012).Article
CAS
PubMed
PubMed Central
Google Scholar
Chacón-Duque, J.-C. et al. Latin Americans show wide-spread Converso ancestry and imprint of local Native ancestry on physical appearance. Nat. Commun. 9, 5388 (2018).Article
ADS
PubMed
PubMed Central
MATH
Google Scholar
Leslie, S. et al. The fine-scale genetic structure of the British population. Nature 519, 309–314 (2015).Article
CAS
PubMed
PubMed Central
MATH
Google Scholar
Cunliffe, B. Facing the Ocean: The Atlantic and Its Peoples 8000 BC-AD 1500 (Oxford Univ. Press, 2001).Taylor, A., Weale, A. & Ford, S. Bronze Age, Iron Age and Roman Landscapes of the Costal Plain, and a Late Iron Age Warrior Burial at North Bersted, Bognor Regis, West Sussex (Thames Valley Archaeological Services, 2014).Fischer, C.-E. et al. Origin and mobility of Iron Age Gaulish groups in present-day France revealed through archaeogenomics. iScience 25, 104094 (2022).Article
ADS
PubMed
PubMed Central
MATH
Google Scholar
Ball, M. J. & Müller, N. (eds) The Celtic Languages 2nd edn (Routledge, 2009).Gretzinger, J. et al. Evidence for dynastic succession among early Celtic elites in Central Europe. Nat. Hum. Behav. https://doi.org/10.1038/s41562-024-01888-7 (2024).Article
PubMed
PubMed Central
Google Scholar
Holden, C. J., Sear, R. & Mace, R. Matriliny as daughter-biased investment. Evol. Hum. Behav. 24, 99–112 (2003).Article
Google Scholar
Jones, D. The matrilocal tribe: an organization of demic expansion. Hum. Nat. 22, 177–200 (2011).Article
PubMed
MATH
Google Scholar
Korotayev, A. Form of marriage, sexual division of labor, and postmarital residence in cross-cultural perspective: a reconsideration. J. Anthropol. Res. 59, 69–89 (2003).Article
Google Scholar
Divale, W. T. Migration, external warfare, and matrilocal residence. Behav. Sci. Res. 9, 75–133 (1974).Article
Google Scholar
Ember, M. & Ember, C. R. The conditions favoring matrilocal versus patrilocal residence. Am. Anthropol. 73, 571–594 (1971).Article
Google Scholar
Moravec, J. C., Marsland, S. & Cox, M. P. Warfare induces post-marital residence change. J. Theor. Biol. 474, 52–62 (2019).Article
ADS
PubMed
MATH
Google Scholar
Redfern, R. & Chamberlain, A. Demographic analysis of Maiden Castle hillfort: evidence for conflict in Late Iron Age and Early Roman period. J. Paleopathol. 1, 68–73 (2011).Article
Google Scholar
Waddington, C. Excavations at Fin Cop, Derbyshire: an Iron Age hillfort in conflict? Archaeol. J. 169, 159–236 (2012).Article
MATH
Google Scholar
Smith, M. Mortal Wounds: The Human Skeleton as Evidence for Conflict in the Past (Pen and Sword, 2017).Thorpe, N. in Materialisierung von Konflikten/Materialisation of Conflicts (eds Hansen, S. & Krause, R.) 259–276 (LOEWE-Schwerpunkt Prähistorische Konfliktforschung Universität Frankfurt, 2020).Mattison, S. M., Quinlan, R. J. & Hare, D. The expendable male hypothesis. Philos. Trans. R. Soc. Lond. B 374, 20180080 (2019).Article
MATH
Google Scholar
Robinson, A. L. & Gottlieb, J. How to close the gender gap in political participation: lessons from matrilineal societies in Africa. Br. J. Polit. Sci. 51, 68–92 (2021).Article
MATH
Google Scholar
Lowes, S. Kinship structure & women: evidence from economics. Daedalus 149, 119–133 (2020).Article
MATH
Google Scholar
Russell, M. et al. The Durotriges Project, phase one: an interim statement. Proc. Dorset Nat. Hist. Archeol. Soc. 135, 217–221 (2014).MATH
Google Scholar
Russell, M. et al. The Durotriges Project, phase two: an interim statement. Proc. Dorset Nat. Hist. Archeol. Soc. 136, 157–161 (2015).MATH
Google Scholar
Russell, M. et al. The Durotriges Project, phase three: an interim statement. Proc. Dorset Nat. Hist. Archeol. Soc. 137, 173–177 (2016).MATH
Google Scholar
Yang, D. Y., Eng, B., Waye, J. S., Dudar, J. C. & Saunders, S. R. Technical note: improved DNA extraction from ancient bones using silica-based spin columns. Am. J. Phys. Anthropol. 105, 539–543 (1998).Article
CAS
PubMed
Google Scholar
Gamba, C. et al. Genome flux and stasis in a five millennium transect of European prehistory. Nat. Commun. 5, 5257 (2014).Article
ADS
CAS
PubMed
MATH
Google Scholar
Boessenkool, S. et al. Combining bleach and mild predigestion improves ancient DNA recovery from bones. Mol. Ecol. Resour. 17, 742–751 (2017).Article
CAS
PubMed
Google Scholar
Dabney, J. & Meyer, M. Extraction of highly degraded DNA from ancient bones and teeth. Methods Mol. Biol. 1963, 25–29 (2019).Article
CAS
PubMed
MATH
Google Scholar
Meyer, M. & Kircher, M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010, db.prot5448 (2010).Article
MATH
Google Scholar
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12 (2011).Article
Google Scholar
Schubert, M., Lindgreen, S. & Orlando, L. AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Res. Notes 9, 88 (2016).Article
PubMed
PubMed Central
Google Scholar
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).Article
CAS
PubMed
PubMed Central
MATH
Google Scholar
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).Article
PubMed
PubMed Central
MATH
Google Scholar
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).Article
CAS
PubMed
PubMed Central
MATH
Google Scholar
Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10, giab008 (2021).Article
PubMed
PubMed Central
MATH
Google Scholar
Weissensteiner, H. et al. HaploGrep 2: mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. 44, W58–W63 (2016).Article
CAS
PubMed
PubMed Central
Google Scholar
van Oven, M. & Kayser, M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum. Mutat. 30, E386–E394 (2009).Article
PubMed
MATH
Google Scholar
Nei, M. & Roychoudhury, A. K. Sampling variances of heterozygosity and genetic distance. Genetics 76, 379–390 (1974).Article
MathSciNet
CAS
PubMed
PubMed Central
MATH
Google Scholar
Nei, M. & Tajima, F. DNA polymorphism detectable by restriction endonucleases. Genetics 97, 145–163 (1981).Article
CAS
PubMed
PubMed Central
MATH
Google Scholar
Mathieson, I. et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature 528, 499–503 (2015).Article
ADS
CAS
PubMed
PubMed Central
MATH
Google Scholar
Brunel, S. et al. Ancient genomes from present-day France unveil 7,000 years of its demographic history. Proc. Natl Acad. Sci. USA 117, 12791–12798 (2020).Article
ADS
CAS
PubMed
PubMed Central
MATH
Google Scholar
Dulias, K. et al. Ancient DNA at the edge of the world: continental immigration and the persistence of Neolithic male lineages in Bronze Age Orkney. Proc. Natl Acad. Sci. USA 119, e2108001119 (2022).Article
CAS
PubMed
PubMed Central
Google Scholar
Margaryan, A. et al. Population genomics of the Viking world. Nature 585, 390–396 (2020).Article
ADS
CAS
PubMed
MATH
Google Scholar
Allentoft, M. E. et al. Population genomics of Bronze Age Eurasia. Nature 522, 167–172 (2015).Article
ADS
CAS
PubMed
MATH
Google Scholar
Damgaard, P. et al. 137 ancient human genomes from across the Eurasian steppes. Nature 557, 369–374 (2018).Article
ADS
CAS
PubMed
MATH
Google Scholar
International Multiple Sclerosis Genetics Consortium & The Wellcome Trust Case Control Consortium 2. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 476, 214–219 (2011).Article
ADS
Google Scholar
Patterson, N., Price, A. L. & Reich, D. Population structure and eigenanalysis. PLoS Genet. 2, e190 (2006).Article
PubMed
PubMed Central
MATH
Google Scholar
Patterson, N. et al. Ancient admixture in human history. Genetics 192, 1065–1093 (2012).Article
PubMed
PubMed Central
MATH
Google Scholar
Broushaki, F. et al. Early Neolithic genomes from the eastern Fertile Crescent. Science 353, 499–503 (2016).Article
ADS
CAS
PubMed
PubMed Central
Google Scholar
Lazaridis, I. et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513, 409–413 (2014).Article
ADS
CAS
PubMed
PubMed Central
MATH
Google Scholar
Brace, S. et al. Ancient genomes indicate population replacement in Early Neolithic Britain. Nat. Ecol. Evol. 3, 765–771 (2019).Article
ADS
PubMed
PubMed Central
MATH
Google Scholar
Cassidy, L. M. et al. A dynastic elite in monumental Neolithic society. Nature 582, 384–388 (2020).Article
ADS
CAS
PubMed
PubMed Central
MATH
Google Scholar
Yaka, R. et al. Variable kinship patterns in Neolithic Anatolia revealed by ancient genomes. Curr. Biol. 31, 2455–2468 (2021).Article
CAS
PubMed
PubMed Central
MATH
Google Scholar
Jones, E. R. et al. The Neolithic transition in the Baltic was not driven by admixture with Early European Farmers. Curr. Biol. 27, 576–582 (2017).Article
CAS
PubMed
PubMed Central
MATH
Google Scholar
González-Fortes, G. et al. Paleogenomic evidence for multi-generational mixing between Neolithic farmers and Mesolithic hunter-gatherers in the Lower Danube Basin. Curr. Biol. 27, 1801–1810 (2017).Article
PubMed
PubMed Central
MATH
Google Scholar
Mallick, S. et al. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature 538, 201–206 (2016).Article
ADS
CAS
PubMed
PubMed Central
MATH
Google Scholar
Rubinacci, S., Ribeiro, D. M., Hofmeister, R. J. & Delaneau, O. Efficient phasing and imputation of low-coverage sequencing data using large reference panels. Nat. Genet. 53, 120–126 (2021).Article
CAS
PubMed
Google Scholar
1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 526, 68–74 (2015).Article
Google Scholar
Browning, B. L., Tian, X., Zhou, Y. & Browning, S. R. Fast two-stage phasing of large-scale sequence data. Am. J. Hum. Genet. 108, 1880–1890 (2021).Article
CAS
PubMed
PubMed Central
MATH
Google Scholar
Traag, V. A., Waltman, L. & van Eck, N. J. From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. 9, 5233 (2019).Article
ADS
CAS
PubMed
PubMed Central
MATH
Google Scholar
Kharchenko, P., Petukhov, V., Wang, Y. & Biederstedt, E. leidenAlg: implements the Leiden algorithm via an R interface. GitHub https://github.com/kharchenkolab/leidenAlg (2023).Schliep, K. P. phangorn: phylogenetic analysis in R. Bioinformatics 27, 592–593 (2011).Article
CAS
PubMed
MATH
Google Scholar
Caballero, M. et al. Crossover interference and sex-specific genetic maps shape identical by descent sharing in close relatives. PLoS Genet. 15, e1007979 (2019).Article
PubMed
PubMed Central
MATH
Google Scholar
Antonio, M. L. et al. Ancient Rome: a genetic crossroads of Europe and the Mediterranean. Science 366, 708–714 (2019).Article
ADS
CAS
PubMed
PubMed Central
MATH
Google Scholar
Fernandes, D. M. et al. A genomic Neolithic time transect of hunter-farmer admixture in central Poland. Sci. Rep. 8, 14879 (2018).Article
ADS
CAS
PubMed
PubMed Central
MATH
Google Scholar
Freilich, S. et al. Reconstructing genetic histories and social organisation in Neolithic and Bronze Age Croatia. Sci. Rep. 11, 16729 (2021).Article
ADS
CAS
PubMed
PubMed Central
MATH
Google Scholar
Gretzinger, J. et al. The Anglo-Saxon migration and the formation of the early English gene pool. Nature 610, 112–119 (2022).Article
ADS
CAS
PubMed
PubMed Central
Google Scholar
Olalde, I. et al. The Beaker phenomenon and the genomic transformation of northwest Europe. Nature 555, 190–196 (2018).Article
ADS
CAS
PubMed
PubMed Central
MATH
Google Scholar
Seguin-Orlando, A. et al. Heterogeneous hunter-gatherer and steppe-related ancestries in Late Neolithic and Bell Beaker genomes from present-day France. Curr. Biol. 31, 1072–1083 (2021).Article
CAS
PubMed
Google Scholar
Žegarac, A. et al. Ancient genomes provide insights into family structure and the heredity of social status in the early Bronze Age of southeastern Europe. Sci. Rep. 11, 10072 (2021).Article
ADS
PubMed
PubMed Central
MATH
Google Scholar
Mathieson, I. et al. The genomic history of southeastern Europe. Nature 555, 197–203 (2018).Article
ADS
CAS
PubMed
PubMed Central
MATH
Google Scholar
Delaneau, O., Zagury, J.-F. & Marchini, J. Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods 10, 5–6 (2013).Article
CAS
PubMed
Google Scholar
Download referencesThe Durotriges Project is the cumulative product of a large team of Bournemouth University staff, students and volunteers, and we acknowledge the input of all involved since 2009. Special thanks must go to our hosts, R. Hill and family, for generously permitting and facilitating all aspects of archaeological fieldwork. This work was funded by Science Foundation Ireland/Health Research Board/Wellcome Trust Biomedical Research Partnership Investigator Award no. 205072 to D.G.B., ‘Ancient Genomics and the Atlantic Burden’, and a Taighde Éireann – Research Ireland Laureate Award (IRCLA/2022/126) to L.M.C., ‘Ancient Isle’. We thank E. Kenny and the team at Trinseq (Trinity College Dublin) for sequencing support.Department of Genetics, Trinity College Dublin, Dublin, IrelandLara M. Cassidy, Valeria Mattiangeli, Emily M. Breslin, Iseult Jackson, Maeve McCann, Harry Little, Ciarán G. O’Connor, Beth Heaslip & Daniel G. BradleyDepartment of Archaeology and Anthropology, Bournemouth University, Bournemouth, UKMiles Russell, Martin Smith, Gabrielle Delbarre & Paul CheethamDepartment of Life and Environmental Sciences, Bournemouth University, Bournemouth, UKHarry ManleySchool of Mathematics, University of Bristol, Bristol, UKDaniel LawsonDepartment of Genomics, University of Tartu, Tartu, EstoniaPhillip EndicottDepartment of Linguistics, University of Hawai‘i at Mānoa, Mānoa, HI, USAPhillip EndicottDFG Center for Advanced Studies, University of Tübingen, Tübingen, GermanyPhillip EndicottÉco-anthropologie, Musée de l’Homme, Paris, FrancePhillip EndicottYou can also search for this author in
PubMed Google ScholarYou can also search for this author in
PubMed Google ScholarYou can also search for this author in
PubMed Google ScholarYou can also search for this author in
PubMed Google ScholarYou can also search for this author in
PubMed Google ScholarYou can also search for this author in
PubMed Google ScholarYou can also search for this author in
PubMed Google ScholarYou can also search for this author in
PubMed Google ScholarYou can also search for this author in
PubMed Google ScholarYou can also search for this author in
PubMed Google ScholarYou can also search for this author in
PubMed Google ScholarYou can also search for this author in
PubMed Google ScholarYou can also search for this author in
PubMed Google ScholarYou can also search for this author in
PubMed Google ScholarYou can also search for this author in
PubMed Google ScholarYou can also search for this author in
PubMed Google ScholarThis study was designed by L.M.C., D.G.B. and P.E. L.M.C., V.M., E.M.B., I.J., M.M., H.L., C.G.O’C. and B.H. performed laboratory work. L.M.C. processed and analysed the data, with contributions from D.L. M.R., G.D., M.S., P.C. and H.M. gave access to samples, provided archaeological and osteological context, and aided in the interpretation of results. L.M.C. and D.G.B. co-wrote the manuscript with considerable input from P.E., M.R., M.S., P.C. and D.L.Correspondence to
Lara M. Cassidy.The authors declare no competing interests.Nature thanks the anonymous reviewers for their contribution to the peer review of this work.Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.Only sites with a sample size of six or higher are considered and first-degree relative pairs are removed. Of the 23 sites considered, 11 have a h value of 1 (no haplotype represented more than once). These are marked as black points on the map and are composed almost entirely of genetically unrelated individuals. Pie charts show haplogroup frequencies for the remaining 12 sites. Haplogroups with a count above one are emphasised with a black outline in the pie charts. Dotted lines are used to split haplogroups into downstream subclades based on additional mutations. Iron Age tribe names recorded by classical writers and their approximate geographic locations are shown. The haplotypes of all British Iron Age samples used in this analysis are given in Supplementary Table 12.The bottom panel shows median positions of modern European populations (grey text with first three letters of country), Iron Age continental populations (black circles with first three letters of country) and Iron Age British and coastal French populations (coloured circles). Iron Age continental French genomes have been split into northern (nFr) and southern (sFr) populations. The map provides a colour key for British and coastal French Iron Age populations (Supplementary Table 12). Sites with data for more than one individual are shown as large circles with two letter identifiers. The median values for these multi-individual sites are plotted in the top PCA panel. The grey box delimits the area within two standard deviations of the mean value for the British Iron Age population along PC1 and PC2. Outlying individuals that fall beyond this area are plotted using their two-letter site ID. Two outliers from the current study are plotted using their full IDs (WBK02 and WBK30). Two data points from Urville-Nacqueville41 on the Normandy coast are also highlighted (brown). One of these (UN19) places further away from French populations, in agreement with SOURCEFIND results.Surrogate and target individuals are listed with their population IDs in Supplementary Table 12. Raw values are available in Supplementary Table 17. The top rows show averaged profiles for Bronze and Iron Age archaeological sites in Britain, France and the Netherlands. British site profiles are outlined in black if the site contains one or more individual outliers (those possessing a level of British Early Bronze Age ancestry two standard deviations below the population mean). The bottom row shows ancestry profiles for these outlying genomes. We note that all Middle and Late Iron Age outliers derive from the channel core region. We identify two previously reported17 EEF outliers from Margetts Pit and Cliffs End Farm in Middle-Late Bronze Age Kent, who possess little to no British Early Bronze Age ancestry. (BA: Bronze Age, EIA: Early Iron Age, MIA: Middle Iron Age, LIA: Late Iron Age).Genetic relatives are defined as individuals who share ≥ 24 cM across at least three IBD segments that are 4 cM or longer (Supplementary Note 5.2). a, This shows the frequency of relative pairs identified within archaeological sites, where a value of one indicates that all pairs are relatives and a value of zero indicates no pairs are relatives. b, This shows archaeological sites that are linked by genetic relatives (Supplementary Table 14; common colours denote a set of linked sites). Most pairs of relatives are identified between neighbouring sites (2–40 km), but several long-distance relative pairs are also identified. An individual from Winterborne Kingston (WBK01) has two relatives at Carsington Pasture Cave in the midlands (red). Three Scottish samples from northern coastal sites also show a relationship (green).The normalised amounts of IBD sharing (cM) between 12 sites in northern England. Each panel shows pairwise values for a specific site (marked with an X) and the other 11. Sites highlighted in red text are all located west of the River Derwent (highlighted in white) and show excessive IBD sharing with one another. The Roman burials of Driffield Terrace18 show low amounts of IBD sharing with sites across the region, likely due to the individuals buried there being non-local in origin.For each measure, we calculate the average total amount of IBD shared (cM), represented using a colour scale. For within genome (i.e. runs of homozygosity) and within site comparisons we also show the average size of shared IBD segments. Within region sharing is calculated as the average amount of IBD shared (cM) between sites within a geographic region, defined based on present-day district divisions (see Supplementary Note 5.4, Supplementary Table 12). It is a more robust measure of relative population sizes than within site and within genome IBD sharing, as these latter can be confounded by kinship and cultural practices such as consanguineous marriage. We see lowest values for within region IBD sharing in France and the southeast of England. For within genome IBD sharing, we take the population average of the total length of runs of homozygosity for each archaeological site. If the average total length is above 3 cM we plot the site in the bottom panel. In Britain, sites with reduced runs of homozygosity (top panel) are concentrated in south central and southeastern regions indicative of larger population sizes. Within site IBD sharing (average length shared between individuals in a site) is plotted in a similar fashion.The percentage of EEF ancestry is shown on the y axis, estimated with qpAdm following the approach of Patterson et al. (2022)17. The map shows the location of Bronze and Iron Age sites used for analysis. No genomes post-dating AD 250 were included. Samples are grouped by geography, with the “channel core” zone south of the River Thames subdivided into western (dark red), central (red) and eastern (yellow) regions. On the right-hand side, EEF ancestry is plotted against the date estimate for each genome and a rolling average line is shown (window size: 500 years, step size: 50 years). On the left-hand side, boxplots (Tukey’s method) show the spread of values for different 500-year time bins for each region. These bins are demarcated with grey dashed lines on the rolling average plot. Time bin date ranges are labelled with respect to the approximate archaeological period they centre on (EBA: Earlier Bronze Age, MBA: Middle Bronze Age, LBA: Later Bronze Age, EIA: Earlier Iron Age, LIA: Later Iron Age). Significant changes (Welch’s t-test, two-tailed; p < 0.05) between bins (n > 4) are highlighted with arrows. The period between 1000-875 BC is highlighted with a grey rectangle in the rolling average plots. This period has been previously associated with a population-wide increase in EEF ancestry in southern Britain17.Surrogate and target individuals are listed with their population IDs in Supplementary Table 12. Individual ancestry profiles for the French site of Urville-Nacqueville are also shown41, two of which (white outline) are below the coverage threshold to which the remainder of the dataset was subject. They are included here to demonstrate that the individual (UN19) with high levels of British Early Bronze Age ancestry is likely an outlier at the site, in agreement with the projection PCA analysis (Extended Data Fig. 2). Note that the French Iron Age samples used as a surrogate population are 1240k SNP capture data, while French target populations on the map are whole genome shotgun sequence data.Supplementary Notes 1–6, Figs. 1–35 and Tables 19–25.Supplementary Tables 1–18.Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.Reprints and permissionsCassidy, L.M., Russell, M., Smith, M. et al. Continental influx and pervasive matrilocality in Iron Age Britain.
Nature (2025). https://doi.org/10.1038/s41586-024-08409-6Download citationReceived: 07 May 2024Accepted: 14 November 2024Published: 15 January 2025DOI: https://doi.org/10.1038/s41586-024-08409-6Anyone you share the following link with will be able to read this content:Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
Advertisement
Nature (Nature)
ISSN 1476-4687 (online)
ISSN 0028-0836 (print)
© 2025 Springer Nature LimitedSign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.